Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data
This repository is intended to support reproducibility of our recent manuscript: “Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data.” The complete text and supplementary materials of the preprint are made freely available via bioRxiv.[1]
To enable rapid reproduction of the published figures, we have made a set of pre-assembled Seurat objects available as part of the R package “BatchNorm”, available on github.
Installation
To install the package (along with select published datasets in support of the manuscript) in R, run:
if(!requireNamespace("devtools", quietly = TRUE)) {
install.packages("devtools")
}
devtools::install_github("Ghosn-Lab/BatchNorm")The package should install within a few minutes, and all functions can be reproduced on a standard desktop or laptop without special hardware.
Examples
To reproduce the results and figures as presented in the manuscript, you can follow along with our vignettes here
Biaxial Gating of a Single Sample
The first vignette describes exactly how cells from a single sample/dataset are categorized as part of a multi-dataset workflow. Following along with this guide will exactly reproduce figure S3 [1].
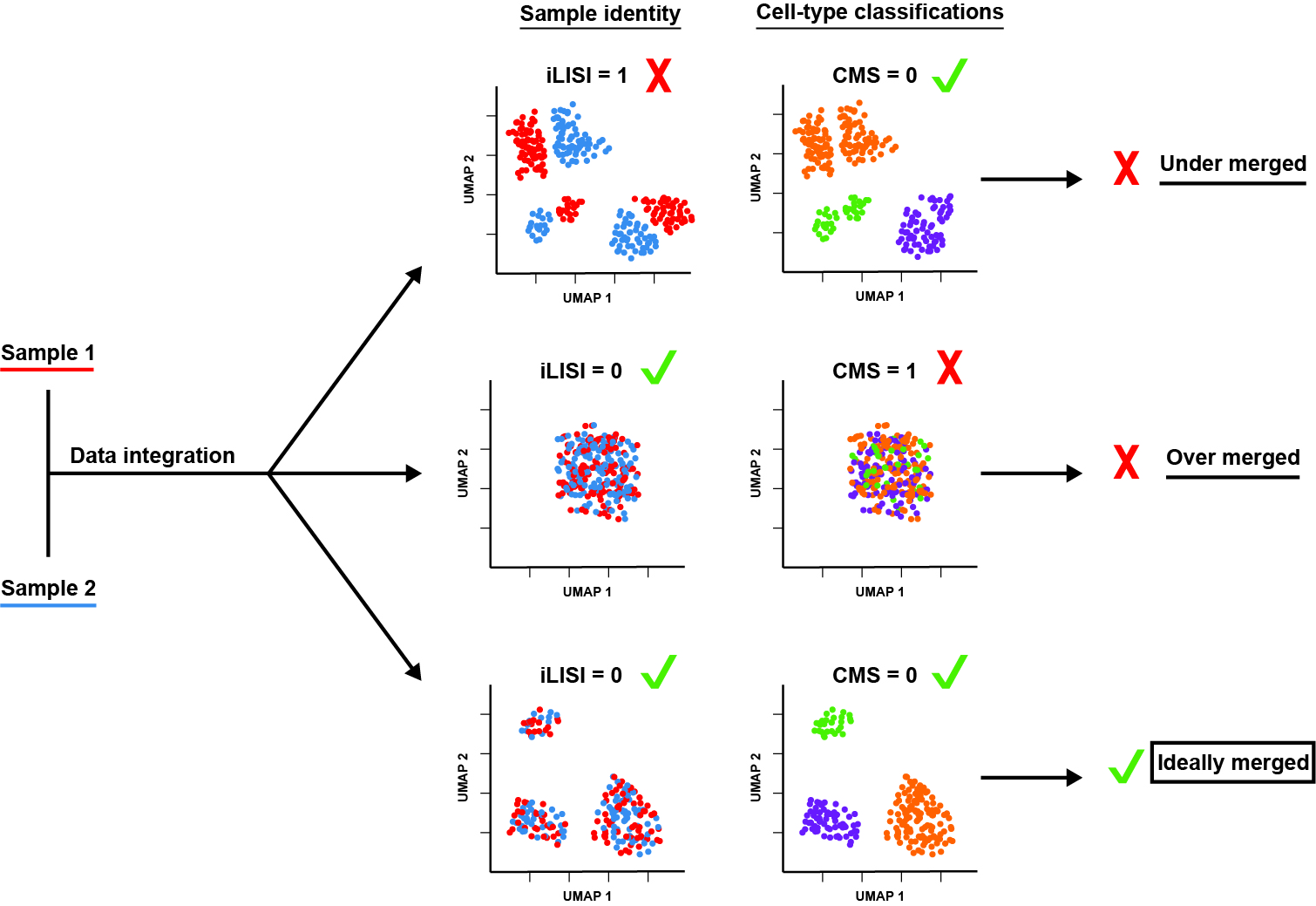
Batch effects are generated by sample donor
The second vignette describes how we determine the batch effects of three identically prepared and sequenced PBMC datasets, using CMS and iLISI scoring. The results of this workflow are depicted in figure 2A [1].
Batch effects are generated by sequencing depth, but not sequencing replicates alone
The third vignette describes how we use CMS and iLISI scoring to determine the batch effects occuring from identical sequencing at different times, and from sequencing depth. The sample duplicating scheme is depicted in figure 2B, while results of this workflow are depicted in figures 2C-D [1].
Pooling samples for sequencing does not appreciably improve the measured batch effect
The fourth vignette depicts the analysis of scRNA-Seq libraries from two different donors, sequenced to similar depth (Samples 4-A, 4-B and 5-A) were either sequenced at the same time (“sequenced together” samples 4-A & 5-A) or at different times (“sequenced separately” samples 4-B & 5-A). The full results of this test are described in our published Figure 3 [1].
Common batch-effect correction methods imperfectly resolve batch effect
The fifth vignette applies three popular batch effect correction workflows to scRNA-Seq libraries from three different donors (the batch-specific signatures of which are established and detailed in Figure 2 and the vignette titled “Sample Donor Effects”). We assess the performance of each method using CMS and iLISI scoring. The full results of this test are described in our published Figure 4 and accompanying text [1].
Data normalization and merging strategies differentially impact the measured batch effect
The sixth vignette details how we measure the CMS and iLISI scores resulting from two data normalization/scaling methods (log-normalization + scaling vs. SCTransform). We perform each method twice - either on individual samples (prior to merging) or on a joint object of all samples (after merging). The results of this analysis are presented in greater detail in Figure 5 of our manuscript [1].
References
[1] Babcock, B. R. et al.
Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data. bioRxiv (2021) DOI: https://doi.org/10.1101/2021.08.18.456898